New Neuroglancer feature: import annotations from CSV files
Annotations are a useful feature of Neuroglancer. In previous posts, we’ve gone over how to add annotations to mark regions of interest in our lightsheet volumes. In the Brain Registration and Histology Core Facility at PNI, we also frequently use annotations to display detected cell locations. In cases like this, where we want to create lots of annotations and we already know their locations in our volume, it is very tedious to manually add them. The goal of this new feature is to provide a more convenient way to upload many annotations simultaneously.
This is now possible using the “Upload from CSV” button. This screenshot shows the required format of the uploadable CSV file.
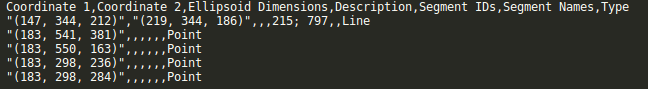
The video below explains the formatting of this file as well as some other features of the new import csv button.
Note that importing annotations from a CSV file is not efficient for very large numbers of annotations. If you start noticing that loading your CSV file is taking a while (usually for ~10^4 or more annotations), you may want to consider a second method for getting the annotations into Neuroglancer. This other method involves creating a precomputed annotation layer and then hosting the layer. This method can load up to 10^7 annotations in less than a second. This method is a bit more work and is not covered in this post, but it will be covered in a future post. Feel free to reach out in the meantime if you want to explore this method.